Where does transcription occur and where does translation occur in the cell?
Transcription occurs in the nucleus, whereas translation occurs in the
cytoplasm.
Explanation:
Terms transcriptionand translation in biology are generally related to the DNA and its
properties. Human cells replicate. In order to do that, they have to
produce the same constituents for the new cell that is to be made. The only
way to do that is by producing proteins. The proteins are produced in the
process that is called protein synthesis. The first step is in the nucleus
where a particular gene is being expressed so it enables all protein
factors to come and to replicate that part of a chromosome. This is
finished when mRNA, the same single-stranded genetic code of a particular
gene, is formed. This is transcription.Right after that, the mRNA enters
the cytoplasm through nuclear pores. There, it could be translated into
proteins by ribosomes. This process is called translation.
While discussing transcription and translation, there is confusion where does transcription occur and where does the translation occurs. Transcription is the process by which a message from DNA is copied to messenger RNA and this process is completed in the cell nucleus. The translation is the process of decrypting the information present in mRNA and takes place on ribosomes, in the cytoplasm of the cell.

What is Transcription?
Transcription is a process during which a specific DNA segment is copied (transcribed) into messenger RNA (mRNA) and the process is regulated by an enzyme called RNA polymerase.
It takes place in the following steps:
1. Initiation
During initiation double strand of DNA is uncoiled by the action of RNA polymerase and each strand is copied into mRNA.
2. Elongation
Elongation of RNA is the process by which the addition of nucleotides is being continued to the growing strand of mRNA by the catalyst activity of the polymerase.
3. Termination
During termination phase of transcription, a terminating sequence of nucleotides is being added and the chain is completed.
4. Processing and splicing
mRNA formed is not in a final form. It contains undesirable segments in the chain. By the process of processing and splicing, those segments are removed and the fine form of mRNA is obtained.
Where does transcription occur?
All this transcription process takes place in the nucleus, a membrane bound organelle present in the cell. The nucleus is the most important organelle of the cell and controls all the processes involved in growth and reproduction. After completion of transcription, mRNA moves out of the nucleus, and the rest of the gene expression takes place in the cytoplasm.
What is RNA splicing?
The pre mRNA that is formed from DNA in the previous step also contains some undesirable segments that are not needed in further processes, they are known as introns. Hence, to get rid of these extra segments, RNA is processed and actual mRNA is formed.
This process of chopping of pre mRNA is called splicing.
What is a gene?
Gene is actually a basic hereditary unit that controls which properties will be inherited by offspring and to what extent.
DNA and RNA contain these genes, in other words, they carry genetic information which is further expressed in the form of proteins.
Summary
Transcription is the process by which the nucleotide sequence of DNA is copied into messenger RNA and this process takes place in the nucleus.
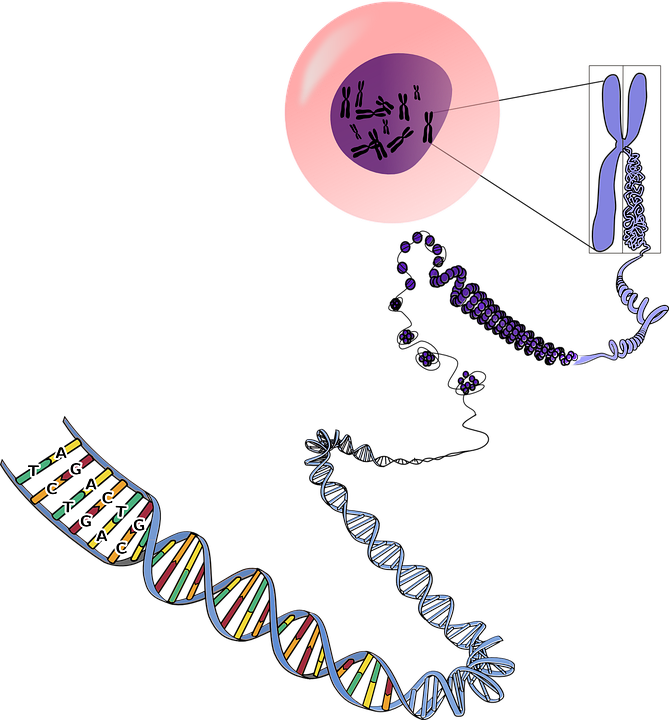
What is translation?
Translation in biology is the process in which the information is carried by mRNA is translated into proteins.
-
The mRNA that is formed by transcription of DNA is now carried out of the nucleus and transferred to the cytoplasm, at the location where protein synthesis machinery is located.
-
This desired location is the ribosome, where the proteins are synthesized according to the information carried by mRNA.
-
However, there is no direct involvement of mRNA in the process of protein manufacturing. Here, another type of RNA is used that is known as tRNA.
-
This tRNA is also located in the cytoplasm and assists in the formation of proteins along with rRNA (ribosomal RNA).
-
The process by which the protein synthesis is carried out with the help of tRNA and rRNA is defined as translation.
Where does translation occur?
It takes place outside the nucleus and inside the cytoplasm. The cytoplasm is a thick solution that covers all the space inside the cell membrane and outside the nuclear membrane. All the organelles are floating in the cytoplasm.
Summary
Translation is the process by which the information carried by mRNA in the form of a genetic codon is decrypted to build the protein subunits called amino acids. This process takes place in the cytoplasm.
Frequently asked questions
In addition to the above details, people often ask several questions regarding transcription and translation. Some of them have been answered below:
1. What are the three differences between transcription and translation?
| Transcription | Translation |
|---|---|
| 1. Transcription produces mRNA | 1. Translation produces amino acids |
| 2. It takes place in the nucleus. | 2. It takes place in the cytoplasm. |
| 3. It is catalyzed by RNA polymerase | 3. It is assisted by rRNA and mRNA. |
2. Why are the transcription and translation important?
Proteins are the basic structural units of the human body. These proteins have produced the process of translation. Transcription converts the hidden information to produce the proteins into mRNA and this information is then decoded at ribosomes by the process of translation. Thus without transcription and translation, the cell can not exist.
3. What is the end result of translation?
The translation is the process by which information carried by messenger RNA is decoded into amino acids at ribosomes within the cell cytoplasm. Amino acids are building blocks of proteins and proteins are essential for all important happenings of the cell, for example, enzymes, that act as catalysts in several cell processes are proteins. Hence, protein formation is the end result of translation.
4. What is the role of genes in inheritance?
Inheritance is the process by which the properties are transferred from parents to children. These properties are transferred by genes that are the basic hereditary unit and contains all information regarding the genetic makeup of offspring. Thus inheritance and gene expression are the processes that go hand in hand.
Conclusion
Transcription is the process of copying the nucleotide sequence from DNA into mRNA in the nucleus. This process is controlled by RNA polymerase. The translation is the process of expressing the information carried by mRNA into nucleic acids in the cytoplasm. Amino acids are the building blocks of proteins.
Related searches
How do you transcribe DNA into mRNA?
What is inheritance and gene expression?
Evolution,a theory, or fact?
Where does transcription occur where does translation occur?
Transcription of DNA occurs in the nucleus. mRNA translation occurs at ribosomes. Transcription of DNA and translation of DNA are part of protein synthesis. Transcription of DNA take place by copying a length of DNA template (gene coding for a polypeptide) to form mRNA (messenger RNA). Translation occurs when mRNA sequence is converted into complementary amino acid sequence with help of tRNA (transfer RNA) molecules and enzymes like peptidyl transferase at the ribosome.
What is transcription?
Transcription is the first step in gene expression, in which information from a gene is use to construct a functional product such as protein.
The goal of transcription is to make a RNA copy of gene’s DNA sequence.
Key points of transcription
- Transcription is the first step in gene expression. Copying a gene’s DNA sequence to make an RNA molecule in first step.
- Enzyme RNA polymerase perform transcription, which link nucleotides to form an RNA strand using DNA strand as a template.
- Transcription has 3 stages, initiation, elongation and termination.
- In eukaryotes RNA molecule must be processed after transcription: they are spliced and have a 5’ cap and poly A tail on their ends.
- Transcription is controlled separately for each gene.
What are RNA polymerases?
The main enzymes involved in transcription is RNA polymerases. Which use single stranded DNA as a template and to a synthesize complementary strands of RNA.
Transcription happens for individual gene
Transcription is controlled individually for each gene (or in bacteria, for small group of genes that are transcribed together). Cells carefully regulates transcription, transcribing just the genes whose products are needed at a moment
What is translation?
Cytoplasm or endoplasmic reticulum synthesize proteins in which ribosomes are present after the process transcription of DNA to RNA in the cell’s nucleus is called Translation. The entire process is called gene expression. Translation also occurs in 3 stages initiation, elongation and termination.
The genetic code
Messenger RNA (mRNA) is decoded in a ribosome, outside the nucleus in the process of translation to produce a specific amino acid chain, or polypeptide. The folding of polypeptide chain occurs and change into an active protein and performs its functions in the cell. The ribosome perform the function of decoding by inducing the binding of complementary tRNA anticodon sequences to mRNA codons. Amino acids of specific type present in tRNA that are chained together into a polypeptide as the mRNA passes through and is “read” by the ribosome.
Translation in prokaryotes and eukaryotes
Cytoplasm in prokaryotes where the large and small subunits of the ribosome bind to the mRNA when translation occurs. While translation occurs in the cytosol or across the membrane of the endoplasmic reticulum in eukaryotes and process called co-translational translocation. The entire ribosome/mRNA complex binds to the outer membrane of the rough endoplasmic reticulum (ER) in co-translational translocation and the new protein is synthesized and released into the endoplasmic reticulum (ER), polypeptide can be stored inside the ER which is newly formed for future vesicle transport and secretion outside the cell, or immediately secreted.
 From DNA to messenger RNA: transcription
From DNA to messenger RNA: transcription 
![]() Transcription is an RNA biosynthesis that, like that of DNA, relies on base complementarity. A strand of DNA serves as a template. Transcription takes place in the nucleus. It consists of copying coded information contained in the DNA molecule into coded information contained in a messenger RNA molecule.
Transcription is an RNA biosynthesis that, like that of DNA, relies on base complementarity. A strand of DNA serves as a template. Transcription takes place in the nucleus. It consists of copying coded information contained in the DNA molecule into coded information contained in a messenger RNA molecule.
![]() Unlike replication, which involves the entire genome at each cycle, the transcription program is not fixed: only small portions of the genome are transcribed at a given time in the life of the cell and these portions vary according to the development, environment etc …
Unlike replication, which involves the entire genome at each cycle, the transcription program is not fixed: only small portions of the genome are transcribed at a given time in the life of the cell and these portions vary according to the development, environment etc …
![]() Transcription, therefore, begins at a precise point of the DNA to end at an equally precise point, the space between the two constitutes a unit of transcription, a concept close to the cistron but not all exactly the same.
Transcription, therefore, begins at a precise point of the DNA to end at an equally precise point, the space between the two constitutes a unit of transcription, a concept close to the cistron but not all exactly the same.
![]() a single strand of DNA is transcribed, ie serves as a model for the polymerization of ribonucleotides. Indeed, a single strand of DNA in a given location has a meaning in terms of protein, which is why we generally write a DNA sequence in the form of a succession of bases from 5 ‘to 3’. In this convention, we actually represent the strand that has the code (coding strand) but it is the other strand (3 ‘- 5’ in our convention) that is transcribed. The result is an RNA molecule whose 5 ‘3’ orientation matches the NH2 - COOH orientation of the protein. The reading of the code (the translation) is done in the same direction as the transcription.
a single strand of DNA is transcribed, ie serves as a model for the polymerization of ribonucleotides. Indeed, a single strand of DNA in a given location has a meaning in terms of protein, which is why we generally write a DNA sequence in the form of a succession of bases from 5 ‘to 3’. In this convention, we actually represent the strand that has the code (coding strand) but it is the other strand (3 ‘- 5’ in our convention) that is transcribed. The result is an RNA molecule whose 5 ‘3’ orientation matches the NH2 - COOH orientation of the protein. The reading of the code (the translation) is done in the same direction as the transcription.
Note: All the transcription product does not correspond to the code of the protein, on the 5 '(upstream) side a guide or “leader” sequence allows the attachment of the messenger RNA to the ribosome.
![]() Transcription is provided by an RNA polymerase that uses single-stranded DNA (as for replication, local denaturation of the DNA molecule is necessary) but it polymerizes ribonucleotides opposite deoxyribonucleotides.
Transcription is provided by an RNA polymerase that uses single-stranded DNA (as for replication, local denaturation of the DNA molecule is necessary) but it polymerizes ribonucleotides opposite deoxyribonucleotides.
Note: Unlike DNA polymerase which only lengthens pre-existing chains, RNA polymerase can make a dinucleotide at the point of initiation of transcription.
![]() Many protein factors are also involved in ensuring initiation, elongation and termination, but they are not the same as those involved in the replication.
Many protein factors are also involved in ensuring initiation, elongation and termination, but they are not the same as those involved in the replication.
 Work of RNA Polymerase
Work of RNA Polymerase 
![]() Transcription begins with the opening and unwinding of a portion of the DNA double helix molecule. As it progresses along with the DNA, the enzymatic complex that constitutes the RNA polymerase incorporates nucleotides present in the cell medium. This incorporation takes place by the complementarity of the nitrogenous bases with one of the strands of the DNA molecule: adenine (A) is placed opposite thymine (T) and uracil (U) is placed opposite adenine (A), cytosine (C) is placed opposite guanine (G) and vice versa.
Transcription begins with the opening and unwinding of a portion of the DNA double helix molecule. As it progresses along with the DNA, the enzymatic complex that constitutes the RNA polymerase incorporates nucleotides present in the cell medium. This incorporation takes place by the complementarity of the nitrogenous bases with one of the strands of the DNA molecule: adenine (A) is placed opposite thymine (T) and uracil (U) is placed opposite adenine (A), cytosine (C) is placed opposite guanine (G) and vice versa.
![]() The strand of messenger RNA thus synthesized is complementary to the strand of transcribed DNA. The information contained in the messenger RNA is identical to that of the non-transcribed DNA strand. The uracil nucleotide (U) occupies the place of the thymine nucleotide (T) in DNA in RNA.
The strand of messenger RNA thus synthesized is complementary to the strand of transcribed DNA. The information contained in the messenger RNA is identical to that of the non-transcribed DNA strand. The uracil nucleotide (U) occupies the place of the thymine nucleotide (T) in DNA in RNA.
![]() RNA polymerase always moves in the same direction on one strand and in the opposite direction on the other strand. It is detached at a site with particular characteristics signalling the end of the gene. Then, when the RNA polymerase is detached, the two DNA strands join together again as the enzyme progresses, and find themselves as they were before RNA synthesis.
RNA polymerase always moves in the same direction on one strand and in the opposite direction on the other strand. It is detached at a site with particular characteristics signalling the end of the gene. Then, when the RNA polymerase is detached, the two DNA strands join together again as the enzyme progresses, and find themselves as they were before RNA synthesis.
An amplification process
RNA is produced very quickly and in large quantities: several RNA polymerase molecules simultaneously transcribe the same gene.
Several genes can be transcribed simultaneously in the nucleus of the same cell.
 Steps of Transcription
Steps of Transcription 
 Pre-initiation
Pre-initiation
![]() The promoter is a sequence of around one hundred nucleotides located in the regulatory region and designating the start of transcription. It is located upstream from the initiation site and carries sequence elements recognized by RNA polymerase and determining the direction of transcription.
The promoter is a sequence of around one hundred nucleotides located in the regulatory region and designating the start of transcription. It is located upstream from the initiation site and carries sequence elements recognized by RNA polymerase and determining the direction of transcription.
![]() The promoter is made up of short sequences conserved from one transcription unit to another and called consensus sequences
The promoter is made up of short sequences conserved from one transcription unit to another and called consensus sequences
![]() The intrinsic strength (or efficiency) of a promoter is defined by the relative number of initiation per unit time (rate of transcription). It depends on the proportion of the A-T base pairs relative to the CG base pairs, on the position of the sequences at -35 and at -10, in fact, the closer the consensus sequences are to the initiation site, the more the promoter will be. strong and ultimately strong RNA polymerase interaction with the promoter.
The intrinsic strength (or efficiency) of a promoter is defined by the relative number of initiation per unit time (rate of transcription). It depends on the proportion of the A-T base pairs relative to the CG base pairs, on the position of the sequences at -35 and at -10, in fact, the closer the consensus sequences are to the initiation site, the more the promoter will be. strong and ultimately strong RNA polymerase interaction with the promoter.
![]() The promoter acts on the transcription of the DNA segment which is adjacent to it on the same chromosome, the promoter is said to be “cis” active. The promoter is not active on the coding sequences located elsewhere on the chromosome, in this case, we would then speak of the qualifier “ trans ”. The cis is opposed to the trans.
The promoter acts on the transcription of the DNA segment which is adjacent to it on the same chromosome, the promoter is said to be “cis” active. The promoter is not active on the coding sequences located elsewhere on the chromosome, in this case, we would then speak of the qualifier “ trans ”. The cis is opposed to the trans.
![]() The affinity of RNA polymerase for DNA depends on the form of the enzyme: the core enzyme has a weak and non-specific affinity, the holoenzyme has a very strong and specific affinity for the promoter. We can note that the sigma σ subunit in the free state does not bind to DNA. The β 'subunit being basic and the DNA being acidic, it will be this which will facilitate the interaction of the complex with the promoter.
The affinity of RNA polymerase for DNA depends on the form of the enzyme: the core enzyme has a weak and non-specific affinity, the holoenzyme has a very strong and specific affinity for the promoter. We can note that the sigma σ subunit in the free state does not bind to DNA. The β 'subunit being basic and the DNA being acidic, it will be this which will facilitate the interaction of the complex with the promoter.
![]() The sigma σ subunit, therefore, allows specific recognition of the promoter by RNA polymerase and decreases the affinity of the enzyme for non-promoter regions. It acts cyclically, indeed after the initiation made, the sigma factor is detached to be recycled and reused for other initiations of genes.
The sigma σ subunit, therefore, allows specific recognition of the promoter by RNA polymerase and decreases the affinity of the enzyme for non-promoter regions. It acts cyclically, indeed after the initiation made, the sigma factor is detached to be recycled and reused for other initiations of genes.
![]() The RNA polymerase causes the denaturation of the two strands of DNA on 14 pairs of nucleotides, we speak of an open complex that further increases the affinity of the enzyme for the double helix.
The RNA polymerase causes the denaturation of the two strands of DNA on 14 pairs of nucleotides, we speak of an open complex that further increases the affinity of the enzyme for the double helix.
 Initiation
Initiation
![]() Initiation corresponds to the synthesis of the first phosphodiester bond produced by the β subunit which corresponds to the catalytic subunit of RNA polymerase.
Initiation corresponds to the synthesis of the first phosphodiester bond produced by the β subunit which corresponds to the catalytic subunit of RNA polymerase.
![]() The interaction of this subunit is inhibited by rifampicin which thus irreversibly inhibits DNA transcription, as is the case with tuberculosis. A mutation in SU β induces the appearance of bacterial strains resistant to rifampicin.
The interaction of this subunit is inhibited by rifampicin which thus irreversibly inhibits DNA transcription, as is the case with tuberculosis. A mutation in SU β induces the appearance of bacterial strains resistant to rifampicin.
![]() The flow of the first stages of transcription is, therefore:
The flow of the first stages of transcription is, therefore:
- nonspecific binding of the holoenzyme.
- formation of a closed complex at the promoter level
- formation of the open complex (unwinding over 14 nucleotides)
- Placement of the first nucleotide (very often A or G)
- Elongation of 4 to 5 nucleotides
- Detachment of the sigma factor, after the transcription of the first 4-5 nucleotides.
 Elongation
Elongation
![]() Elongation corresponds to the displacement of the transcription bubble along the DNA molecule. The mismatched region is then 70 base pairs. During transcription, the RNA forms a short pairing with the matrix strand of DNA forming a hybrid DNA-RNA helix on about ten base pairs.
Elongation corresponds to the displacement of the transcription bubble along the DNA molecule. The mismatched region is then 70 base pairs. During transcription, the RNA forms a short pairing with the matrix strand of DNA forming a hybrid DNA-RNA helix on about ten base pairs.
![]() Elongation is inhibited by aminoglycosides or amino glucosides.
Elongation is inhibited by aminoglycosides or amino glucosides.
 Termination
Termination
![]() Termination occurs when the enzyme arrives at a specific sequence called a terminator.
Termination occurs when the enzyme arrives at a specific sequence called a terminator.
![]() The terminator is presented in the form of a palindrome (cf. lexicon) which can be perfect or imperfect. This palindrome results in sequence complementarity at the mRNA level which allows the establishment of a hairpin (or stem-loop) structure which is an intra-chain pairing that destabilizes the RNA polymerase up to dissociation.
The terminator is presented in the form of a palindrome (cf. lexicon) which can be perfect or imperfect. This palindrome results in sequence complementarity at the mRNA level which allows the establishment of a hairpin (or stem-loop) structure which is an intra-chain pairing that destabilizes the RNA polymerase up to dissociation.
![]() It can be facilitated by a factor rho ρ depending on the sequence of the terminator, we thus highlight independent rho terminators (about 2/3) and rho dependent terminators (about 1/3):
It can be facilitated by a factor rho ρ depending on the sequence of the terminator, we thus highlight independent rho terminators (about 2/3) and rho dependent terminators (about 1/3):
![]() For independent rho terminators, we find a hairpin structure rich in GC base pairs, followed by a poly-U sequence of about 6 nucleotides allowing easier dissociation of the DNA-RNA hybrid.
For independent rho terminators, we find a hairpin structure rich in GC base pairs, followed by a poly-U sequence of about 6 nucleotides allowing easier dissociation of the DNA-RNA hybrid.
For the rho dependent terminators, we find a shorter hairpin structure that is not rich in GC base pairs and which is not followed by a poly-U sequence. There is therefore a need for the rho factor which has an affinity for RNA short of synthesis, traversing it from 5 ‘to 3’ until the RNA polymerase is found. The rho factor is ATP dependent, the hydrolysis of which will allow the dissociation of the complex.
 Maturation of primary transcripts
Maturation of primary transcripts
![]() The primary transcript corresponds to immature RNA which requires processing in the form of cleavages or base modifications. This maturation is not obligatory. After maturation, mature RNA is obtained. We can take a few examples such as the maturation of the primary transcript giving the rRNAs (45S primary transcript) or the tRNAs by ribonucleases, or the maturation of the primary transcript encoding the mRNAs. Generally, there are few modifications for mRNAs; many mRNAs are translated into proteins while they are still being transcribed (Warning: there is no nucleus in prokaryotes).
The primary transcript corresponds to immature RNA which requires processing in the form of cleavages or base modifications. This maturation is not obligatory. After maturation, mature RNA is obtained. We can take a few examples such as the maturation of the primary transcript giving the rRNAs (45S primary transcript) or the tRNAs by ribonucleases, or the maturation of the primary transcript encoding the mRNAs. Generally, there are few modifications for mRNAs; many mRNAs are translated into proteins while they are still being transcribed (Warning: there is no nucleus in prokaryotes).
![]() The primary transcript codes either for a product, we speak of monocistronic RNA or several products, we will then speak of polycistronic RNA ( cf. beginning of the course, generalities ).
The primary transcript codes either for a product, we speak of monocistronic RNA or several products, we will then speak of polycistronic RNA ( cf. beginning of the course, generalities ).
 From RNA to protein: translation
From RNA to protein: translation 
![]() In the cell, translation takes place at the level of ribosomes: organelles made up of ribosomal RNA (rRNA) and proteins. Ribosomes are made up of two distinct subunits see figures 15 a and b, a large subunit and a small subunit, they are visible under an electron microscope.
In the cell, translation takes place at the level of ribosomes: organelles made up of ribosomal RNA (rRNA) and proteins. Ribosomes are made up of two distinct subunits see figures 15 a and b, a large subunit and a small subunit, they are visible under an electron microscope.
![]() The other actors in the translation are the transfer RNAs (tRNAs see FIG. 7) which carry the amino acids and the messenger RNA (mRNA) itself the sequence of which can be cut into codon from the AUG translation initiation codon.
The other actors in the translation are the transfer RNAs (tRNAs see FIG. 7) which carry the amino acids and the messenger RNA (mRNA) itself the sequence of which can be cut into codon from the AUG translation initiation codon.
![]() In eukaryotes, ribosomes are either free in the cytoplasm or linked to the endoplasmic reticulum (this is called agranular endoplasmic reticulum) or to the nuclear membrane and translation takes place in the cytoplasm.
In eukaryotes, ribosomes are either free in the cytoplasm or linked to the endoplasmic reticulum (this is called agranular endoplasmic reticulum) or to the nuclear membrane and translation takes place in the cytoplasm.
![]() In prokaryotes, transcription and translation are simultaneous whereas in eukaryotes translation takes place in the cytoplasm from mature mRNA.
In prokaryotes, transcription and translation are simultaneous whereas in eukaryotes translation takes place in the cytoplasm from mature mRNA.
![]() The messenger RNA produced in the nucleus passes into the cytoplasm to be translated there: this is the synthesis of proteins.
The messenger RNA produced in the nucleus passes into the cytoplasm to be translated there: this is the synthesis of proteins.
 Stages of Translation
Stages of Translation 
1. Initiation : The ribosome , a small cytoplasmic organelle (“protein assembly workshop”) begins reading the gene whose sequence is copied onto messenger RNA. It recognizes a place on the molecule called an initiator codon . Each ribosome is made up of two subunits: the small subunit carries a messenger RNA reading site and the large subunit has a catalytic site. A ribosome therefore behaves like an enzyme characterized by an active site. It catalyzes the amino acid polymerization of the protein that is being formed.
2. Elongation: The relative displacement of the ribosome and of the messenger RNA is accompanied by the progressive lengthening of the polypeptide chain; to each triplet of nucleotides of the messenger RNA corresponds a precise amino acid which is incorporated into the polypeptide chain being formed. The correspondence between triplets of messenger RNA nucleotides and amino acids is carried out according to the principles of the genetic code. The support for each of 20 amino acids is performed through RNA t (transfer RNA) specific part of which is fixed on each codon. peptide bond allows the attachment of each new amino acid (provided by an RNA transfer) to the last amino acid of the chain being extended. The new amino acid arriving in turn temporarily becomes the last.
3. Termination: The ribosome arrives on one of the three “stop” or “nonsense” codons, codon to which no amino acid corresponds. The dissociation between the messenger RNA and the terminated polypeptide chain then takes place.
 The actors of translation
The actors of translation 
The actors of translation are messenger RNA (mRNA), transfer RNA (tRNA), ribosomes, amino acids, amino-acyl tRNA synthetases, Mg 2+, GTP and ATP.
1) Ribosomes
Ribosomes are made up of ribosomal RNA (rRNA) and proteins and are structured in the form of two subunits in either prokaryotes or eukaryotes ( see also general chapter ). Their size is defined in Svedberg units.
- Prokaryotic ribosomes (70S) consist of a small 30S subunit and a large 50S subunit.
- The 30S subunit consists of a 16S rRNA (1541 nucleotides) and 21 proteins.
- The 50S subunit consists of 23S (2904 nucleotides) and 5S (120 nucleotides) rRNAs as well as 32 proteins.
- Eukaryotic ribosomes (the 80S) consist of a small 40S subunit and a large 60S subunit.
- The 40S subunit is made up of an 18S rRNA and 33 proteins.
- The 60S subunit consists of 28S, 5.8S and 5S rRNAs as well as 49 proteins.
Schematic topography of the bacterial ribosome:
The bacterial ribosome has specific sites:
- Site A: (= Amino-Acid or Acceptor site) binding of amino acids.
- Site P: (= Peptide or Donor site) binding off-Met.
- Site E: (= Exit site) exit of the transfer RNA.
- EF-G site: present at the level of the large sub-unit.
- EF-Tu site: present at the level of the small sub-unit.
Attention in eukaryotes the first amino acid is methionine and not the f-Met present in prokaryotes.
The chaining of ribosomes on mRNA forms the polysome, it makes it possible to increase the efficiency of translation. The minimum distance between two ribosomes is 100 nucleotides.
At the level of the ribosomes associated with the endoplasmic reticulum, the proteins undergoing synthesis enter the vesicles of the reticulum directly after the E site ( cf. cell biology course ).
2) tRNAs
a) Structure of tRNAs and iso-acceptor tRNAs:
tRNAs have a 3-leaf clover-shaped secondary structure and an upside-down L-shaped tertiary structure. During the translation mechanism, there is an antiparallel pairing between mRNA and tRNA: codon-anticodon recognition at the level of the anticodon loop.
There is flexibility in the base-pairing at position 3 of the codon and at position 1 of the anticodon, this flexibility is called wobble.
Note: I is a purine base that can pair with U, C and A but not G.
The tRNAs also have an arm of the amino acid which fixes it in 3 '(CCA) on the ribose, it is about a covalent bond: ester bond rich in energy. The amino acids will thus not arrive free on the ribosome but associated with their respective tRNAs. There are 40 to 60 different tRNAs per cell, so there are several different tRNAs for an amino acid, these are called iso-acceptor tRNAs. (Writing: tARN leu 1)
b) Loading the amino acid onto the tRNA:
The formation of the amino-acyl-tRNA complex ( aa-tRNA ) requires an amino acid - specific Amino-acyl-tRNA-synthetase , which must thus recognize all codon forms of this amino acid. Correct loading of tRNA is an important element in translational fidelity.
The amino acid (aa) is first activated and this activation requires energy in the form of ATP to allow the formation of aa-AMP (mixed anhydride bond).
The bond formed between the tRNA and the amino acid is a covalent bond of the carboxy-ester type. Amino-acyl-tRNA-synthetase is 20 in number in the cell, as many as there are amino acids that are taken into account in translation. The complexed amino acid can thus join the chain.
 The specificities of eukaryotic translation
The specificities of eukaryotic translation 
![]() The ribosome is of different size and composed of different ribosomal RNAs although the general structure and activity are comparable.
The ribosome is of different size and composed of different ribosomal RNAs although the general structure and activity are comparable.
![]() The initiator codon is also AUG and it is usually the 1st AUG present on the mRNA. The ATG triplet can be found which will give the AUG codon on the sense DNA at the level of the Kozak sequence (GCC GCC (A / G) CC ATG G).
The initiator codon is also AUG and it is usually the 1st AUG present on the mRNA. The ATG triplet can be found which will give the AUG codon on the sense DNA at the level of the Kozak sequence (GCC GCC (A / G) CC ATG G).
![]() As said previously, in eukaryotes the first amino acid is methionine and not the f-Met present in prokaryotes. Methionine will most often be removed just after the synthesis of the peptide chain.
As said previously, in eukaryotes the first amino acid is methionine and not the f-Met present in prokaryotes. Methionine will most often be removed just after the synthesis of the peptide chain.
![]() The initiation factors are of the eIF (for eukaryotic initiation factor ) type, from eIF1 to eIF6.
The initiation factors are of the eIF (for eukaryotic initiation factor ) type, from eIF1 to eIF6.
![]() The elongation factors are also of the EF type (EF1α, EF1β and EF2).
The elongation factors are also of the EF type (EF1α, EF1β and EF2).
![]() Termination factors are of the eRF (for eukaryotic Releasing Factor ) type.
Termination factors are of the eRF (for eukaryotic Releasing Factor ) type.
 Conclusion
Conclusion 
The expression of the genetic information contained in DNA comprises two stages inside cells:
- transcription leading to the production of a series of ephemeral copies of the gene, the messenger RNAs;
- translation consisting of a decoding of the information contained in the messenger RNAs in the form of an assembly of amino acids. Ribosomes, globular organelles made up of two subunits, are the “assembly workshops” of proteins.
The process in which cells make proteins is called protein synthesis. It actually consists of two processes: transcription and translation. Transcription takes place in the nucleus. It uses DNA as a template to make an RNA molecule. RNA then leaves the nucleus and goes to a ribosome in the cytoplasm, where translation occurs. Translation reads the genetic code in mRNA and makes a protein.
Transcription is the first part of the central dogma of molecular biology: DNA → RNA. It is the transfer of genetic instructions in DNA to messenger RNA (mRNA). During transcription, a strand of mRNA is made that is complementary to a strand of DNA. Figure 1 shows how this occurs.
Transcription uses the sequence of bases in a strand of DNA to make a complementary strand of mRNA. Triplets are groups of three successive nucleotide bases in DNA. Codons are complementary groups of bases in mRNA.
You can also watch this more detailed video about transcription.
Steps of Transcription
Steps of transcription: initiation, elongation, termination
Initiation is the beginning of transcription. It occurs when the enzyme RNA polymerase binds to a region of a gene called the promoter. This signals the DNA to unwind so the enzyme can ‘‘read’’ the bases in one of the DNA strands. The enzyme is now ready to make a strand of mRNA with a complementary sequence of bases.
Elongation is the addition of nucleotides to the mRNA strand. RNA polymerase reads the unwound DNA strand and builds the mRNA molecule, using complementary base pairs. There is a brief time during this process when the newly formed RNA is bound to the unwound DNA. During this process, an adenine (A) in the DNA binds to an uracil (U) in the RNA.
Termination is the ending of transcription, and occurs when RNA polymerase crosses a stop (termination) sequence in the gene. The mRNA strand is complete, and it detaches from DNA.
This video provides a review of these steps. You can stop watching the video at 5:35. (After this point, it discusses translation, which we’ll discuss in the next outcome.) We turn now to transcription in eukaryotes, a much more complex process than in prokaryotes.
In eukaryotes, transcription and translation take place in different cellular compartments: transcription takes place in the membrane-bounded nucleus, whereas translation takes place outside the nucleus in the cytoplasm. In prokaryotes, the two processes are closely coupled (Figure 28.15). Indeed, the translation of bacterial mRNA begins while the transcript is still being synthesized.
The spatial and temporal separation of transcription and translation enables eukaryotes to regulate gene expression in much more intricate ways, contributing to the richness of eukaryotic form and function. Transcription and Translation. These two processes are closely coupled in prokaryotes, whereas they are spacially and temporally separate in eukaryotes. (A) In prokaryotes, the primary transcript serves as mRNA and is used immediately as the template (more…)
A second major difference between prokaryotes and eukaryotes is the extent of RNA processing. Although both prokaryotes and eukaryotes modify tRNA and rRNA, eukaryotes very extensively process nascent RNA destined to become mRNA.
Primary transcripts (pre-mRNA molecules), the products of RNA polymerase action, acquire a cap at their 5′ ends and a poly(A) tail at their 3′ ends. Most importantly, nearly all mRNA precursors in higher eukaryotes are spliced (Section 5.6.1). Introns are precisely excised from primary transcripts, and exons are joined to form mature mRNAs with continuous messages.
Some mRNAs are only a tenth the size of their precursors, which can be as large as 30 kb or more. The pattern of splicing can be regulated in the course of development to generate variations on a theme, such as membrane-bound and secreted forms of antibody molecules. Alternative splicing enlarges the repertoire of proteins in eukaryotes and is a clear illustration of why the proteome is more complex than the genome.
In prokaryotes, RNA is synthesized by a single kind of polymerase. In contrast, the nucleus of a eukaryote contains three types of RNA polymerase differing in template specificity, location in the nucleus, and susceptibility to inhibitors (Table 28.2).
All these polymerases are large proteins, containing from 8 to 14 subunits and having a total molecular mass greater than 500 kd. RNA polymerase I is located in nucleoli, where it transcribes the tandem array of genes for 18S, 5.8S, and 28S ribosomal RNA (Section 29.3.1). The other ribosomal RNA molecule (5S rRNA, Section 29.3.1) and all the transfer RNA molecules (Section 29.1.2) are synthesized by RNA polymerase III, which is located in the nucleoplasm rather than in nucleoli.
RNA polymerase II, which also is located in the nucleoplasm, synthesizes the precursors of messenger RNA as well as several small RNA molecules, such as those of the splicing apparatus (Section 28.3.5). Although all eukaryotic RNA polymerases are homologous to one another and to prokaryotic RNA polymerase, RNA polymerase II contains a unique carboxyl-terminal domain on the 220-kd subunit; this domain is unusual because it contains multiple repeats of an YSPTSPS consensus sequence.
The activities of RNA polymerase II are regulated by phosphorylation on the serine and threonine residues of the carboxyl-terminal domain. Another major distinction among the polymerases lies in their responses to the toxin α-amanitin, a cyclic octapeptide that contains several modified amino acids.
α-Amanitin is produced by the poisonous mushroom Amanita phalloides, which is also called the death cup or the destroying angel (Figure 28.16). More than a hundred deaths result worldwide each year from the ingestion of poisonous mushrooms. α-Amanitin binds very tightly (Kd = 10 nM) to RNA polymerase II and thereby blocks the elongation phase of RNA synthesis.
Higher concentrations of α-amanitin (1 μM) inhibit polymerase III, whereas polymerase I am insensitive to this toxin. This pattern of sensitivity is highly conserved throughout the animal and plant kingdoms. Eukaryotic genes, like their prokaryotic counterparts, require promoters for transcription initiation. Each of the three types of polymerase has distinct promoters. RNA polymerase I transcribes from a single type of promoter, present only in rRNA genes, that encompasses the initiation site. In some genes, RNA polymerase III responds to promoters located in the normal, upstream position; in other genes, it responds to promoters imbedded in the genes, downstream of the initiation site.
Promoters for RNA polymerase II can be simple or complex (Section 28.2.3). As is the case for prokaryotes, promoters are always on the same molecule of DNA as the gene they regulate. Consequently, promoters are referred to as cis-acting elements.
However, promoters are not the only types of cis-acting DNA sequences. Eukaryotes and their viruses also contain enhancers. These DNA sequences, although not promoters themselves, can enormously increase the effectiveness of promoters. Interestingly, the positions of enhancers relative to promoters are not fixed; they can vary substantially. Enhancers play key roles in regulating gene expression in a specific tissue or developmental stage (Section 31.2.4).
The DNA sequences of cis-acting elements are binding sites for proteins called transcription factors. Such a protein is sometimes called a trans-acting factor because it may be encoded by a gene on a DNA molecule other than that containing the gene being regulated. The binding of a transcription factor to its cognate DNA sequence enables the RNA polymerase to locate the proper initiation site. We will continue our investigation of transcription by examining these cis- and trans-acting elements in turn.
Promoters for RNA polymerase II, like those for bacterial polymerases, are located on the 5′ side of the start site for transcription. The results of mutagenesis experiments, footprinting studies, and comparisons of many higher eukaryotic genes have demonstrated the importance of several upstream regions. For most genes transcribed by RNA polymerase II, the most important cis-acting element is called the TATA box on the basis of its consensus sequence (Figure 28.17).
The TATA box is usually centered between positions -30 and -100. Note that the eukaryotic TATA box closely resembles the prokaryotic - 10 sequence (TATAAT) but is farther from the start site. The mutation of a single base in the TATA box markedly impairs promoter activity. Thus, the precise sequence, not just a high content of AT pairs, is essential.
The TATA box is necessary but not sufficient for strong promoter activity. Additional elements are located between -40 and -150. Many promoters contain a CAAT box, and some contain a GC box (Figure 28.18). Constitutive genes (genes that are continuously expressed rather than regulated) tend to have GC boxes in their promoters.
The positions of these upstream sequences vary from one promoter to another, in contrast with the quite constant location of the -35 region in prokaryotes. Another difference is that the CAAT box and the GC box can be effective when present on the template (antisense) strand, unlike the -35 region, which must be present on the coding (sense) strand.
These differences between prokaryotes and eukaryotes reflect fundamentally different mechanisms for the recognition of cis-acting elements. The -10 and -35 sequences in prokaryotic promoters correspond to binding sites for RNA polymerase and its associated σ factor. In contrast, the TATA, CAAT, and GC boxes and other cis-acting elements in eukaryotic promoters are recognized by proteins other than RNA polymerase itself.
Cis-acting elements constitute only part of the puzzle of eukaryotic gene expression. Transcription factors that bind to these elements also are required. For example, RNA polymerase II is guided to the start site by a set of transcription factors known collectively as TFII (TF stands for transcription factor, and II refers to RNA polymerase II).
Individual TFII factors are called TFIIA, TFIIB, and so on. Initiation begins with the binding of TFIID to the TATA box (Figure 28.19). The key initial event is the recognition of the TATA box by the TATA-box-binding protein (TBP), a 30-kd component of the 700-kd TFIID complex.
TBP binds 105 times as tightly to the TATA box as to noncognate sequences; the dissociation constant of the specific complex is approximately 1 nM. TBP is a saddle-shaped protein consisting of two similar domains (Section 7.3.3; Figure 28.20). The TATA box of DNA binds to the concave surface of TBP. This binding induces large conformational changes in the bound DNA. The double helix is substantially unwound to widen its minor groove, enabling it to make extensive contact with the antiparallel β strands on the concave side of TBP.
Hydrophobic interactions are prominent at this interface. Four phenylalanine residues, for example, are intercalated between base pairs of the TATA box. The flexibility of AT-rich sequences is generally exploited here in bending the DNA. Immediately outside the TATA box, classical B-DNA resumes. This complex is distinctly asymmetric. The asymmetry is crucial for specifying a unique start site and ensuring that transcription proceeds unidirectionally.
TBP bound to the TATA box is the heart of the initiation complex. The surface of the TBP saddle provides docking sites for the binding of other components. Additional transcription factors assemble on this nucleus in a defined sequence.
TFIIA is recruited, followed by TFIIB and then TFIIF—an ATP-dependent helicase that initially separates the DNA duplex for the polymerase. Finally, RNA polymerase II and then TFIIE join the other factors to form a complex called the basal transcription apparatus. Sometime in the formation of this complex, the carboxyl-terminal domain of the polymerase is phosphorylated on the serine and threonine residues, a process required for successful initiation.
The importance of the carboxyl-terminal domain is highlighted by the finding that yeast containing mutant polymerase II with fewer than 10 repeats is not viable. Most of the factors are released before the polymerase leaves the promoter and can then participate in another round of initiation.
